Odovtos-International Journal of Dental Sciences (Odovtos-Int. J. Dent. Sc.), Online First, 2025. ISSN: 2215-3411
https://doi.org/10.15517/0zydcq64

https://revistas.ucr.ac.cr/index.php/Odontos
CLINICAL RESEARCH:
Genetic Association Between RFC1 Gene Polymorphism (rs1051266) and Non-Syndromic Cleft Lip and Palate in the South Indian Population: A Case Control Study
Asociación genética entre el polimorfismo del gen RFC1 (rs1051266) y el labio y paladar hendido no sindrómico en la población del sur de la India: un estudio de casos y controles
A. Sumathi Felicita¹ https://orcid.org/0000-0003-2002-0140
Ritya Mary Jibu² https://orcid.org/0000-0001-6618-746X
Vijayashree Priyadharshini Jayaseelan3 https://orcid.org/0000-0001-7884-5466
¹Professor, Department of Orthodontics, Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences, Saveetha University, Chennai- 600077, India.
²Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences, Saveetha University, Chennai, India.
3Chief scientist, Blue Lab - research centre, Saveetha dental college and Hospitals, Saveetha Institute of Medical & Technical Sciences,
Saveetha University, Chennai - 600077, India.
Correspondence to: A.Sumathi Felicita - sumifeli@hotmail.com
Received: 8-III-2025 Accepted: 30-VII-2025
ABSTRACT: To identify whether an association exists between RFC1 gene polymorphism (rs1051266) and the development of non-syndromic cleft lip and palate. Twenty-five patients with non-syndromic cleft lip and palate (NSCL/P) with ages ranging from 1 year to 17 years belonging to both genders (Group 1) and twenty-five patients without cleft lip and palate malformations as controls (Group 2) were included in the study. Genomic DNA was obtained from whole blood drawn from the patients. The region around the RFC1 gene polymorphism was amplified with sequence-specific primers using polymerase chain reaction (PCR). RFLP technique was used to identify the genotype of the patients. The genotype and allele frequency distributions in both groups were determined. Statistical analysis was done with the Chi-square test. In both groups, the frequency of the GA genotype was greater compared to the GG genotype and AA genotype. There was no statistically significant deviation from Hardy Weinberg Equilibrium with a P value of <0.247 and <0.815 in the case and control groups, respectively. The frequency distribution of the dominant genotype GG had an odd ratio of 0.3506 (0.0791-1.5544) and a P value of 0.1678 and the recessive genotype AA had an odd ratio of1.5556 (0.4187-5.7795) and a P value of 0.5094. The distribution of G allele and A allele between the two group had an odd ratio of 0.6169 (0.2798-1.3597)and a P value of 0.2309. Classification of the genotypes based on genetic models such as dominant, recessive, or additive did not present any significant association between the polymorphism marker and NSCL/P. There was no significant association between RFC1 gene polymorphism (rs1051266) and NSCL/P in the South Indian population.
KEYWORDS: RFC1 gene; Polymorphism; NSCL/P; rs1051266.
RESUMEN: Determinar si existe una asociación entre el polimorfismo del gen RFC1 (rs1051266) y el desarrollo de labio y paladar hendido no sindrómico (LPHNS). El estudio incluyó a 25 pacientes con LPHNS (Grupo 1), con edades entre 1 y 17 años, de ambos sexos, y a 25 individuos sin malformaciones de labio o paladar, quienes constituyeron el grupo control (Grupo 2). Se extrajo ADN genómico a partir de muestras de sangre periférica. La región adyacente al polimorfismo del gen RFC1 fue amplificada mediante cebadores específicos de secuencia utilizando la reacción en cadena de la polimerasa (PCR). La genotipificación se realizó mediante la técnica de polimorfismo de longitud de fragmentos de restricción (RFLP). Se determinaron las frecuencias genotípicas y alélicas, y se realizaron comparaciones estadísticas mediante la prueba de Chi-cuadrado. En ambos grupos, el genotipo GA fue más frecuente en comparación con los genotipos GG y AA. No se observaron desviaciones estadísticamente significativas del equilibrio de Hardy-Weinberg en el grupo de casos (p = 0.247) ni en el grupo control (p = 0.815). La distribución de frecuencias del genotipo dominante (GG) arrojó una razón de momios (OR) de 0.3506 (IC 95%: 0.0791-1.5544; p = 0.1678), mientras que el genotipo recesivo (AA) presentó una OR de 1.5556 (IC 95%: 0.4187–5.7795; p = 0.5094). La comparación de las frecuencias alélicas entre grupos no mostró diferencias significativas (alelo G vs. alelo A: OR = 0.6169; IC 95%: 0.2798-1.3597; p = 0.2309). El análisis con modelos genéticos dominantes, recesivos y aditivos no reveló asociaciones significativas entre el polimorfismo rs1051266 y LPHNS. Los hallazgos sugieren que el polimorfismo rs1051266 del gen RFC1 no se asocia de manera significativa con el riesgo de desarrollar LPHNS en la población del sur de la India.
PALABRAS CLAVE: Gen RFC1; Polimorfismo; NSCL/P; rs1051266.
INTRODUCTION
The most prevalent congenital facial malformations at birth are cleft lip and/or palate, which develop when the frontal prominence and maxillary process fail to fuse during embryogenesis. Non-syndromic lip and palate is multifactorial with a combination of genetic and environmental factors that result in the development of the anomaly (1-3). Environmental factors such as alcohol (4), maternal phenylketonuria (5), hyperthermia (6), hydantoin (7), trimethadion (8), and methotrexate (9) are teratogens linked to the development of cleft lip and palate.
Folate is an essential micronutrient, the deficiency of which has been attributed to the development of cleft lip and palate. Tetrahydrofolate (THF), is a form of folate generally found in food, frequently present in cruciferous vegetables, legumes, fruits, and fortified cereals and grains (10). The recommended dietary allowance for folate is 400 mg in normal adults, but during pregnancy and breastfeeding is 500-600 mg (11). It is advised to take folate supplements during pregnancy since inadequate maternal folate consumption has been strongly linked to an increased risk of neural tube abnormalities (12). Impaired cellular methylation including DNA methylation is a key factor associated with neural tube defects and has been related to folate consumption during pregnancy and birth abnormalities (13).
Several genes such as FOLH1, FOLR1, FOLR2, FOLR3, SLC19A1, SLC46A1, SLC25A32, ABCC2 are associated with the folate one carbon mechanism (14). Polymorphism of such genes can alter the folate mechanism resulting in neural tube defects (14). These defects are associated with the development of cleft lip and palate (15,16). Genetic studies have established the association of numerous gene polymorphisms in the development of cleft lip and palate (17). However, there are not many studies on the influence of RFC1 gene polymorphism (rs1051266) in the development of cleft lip and palate in the Indian population.
Reduced folate carrier 1 (RFC1) gene has been suggested to be associated with the development of cleft lip and palate in humans. Reduced folate carrier (RFC1) is a bidirectional transporter characterized by twelve transmembrane domains (18,19). RFC1 preferentially transports reduced folates, such as N5-methyltetrahydrofolate or N5-formyltetrahydrofolate, which are anionic at physiological pH. It acts as a facilitative anion exchanger, transporting anionic folates into the cell through a cotransport system with an influx of folate and efflux of organic phosphates or sulfates (18). The RFC1 gene is responsible for the encoding of a membrane protein. This protein delivers the 5-methyltetrahydrofolate, a metabolically active form of folate to several cells (20). Polymorphisms of RFC1gene have been considered to be responsible for alteration in folate metabolism and maternal folate intake (21). The missense variation rs1051266 (A80G) in exon 2 was proposed as a risk factor for NSCL/P. Hence the present study was proposed to determine if a polymorphism of the RFC1 gene (rs1051266) was responsible for NSCL/P in the South Indian population. The null hypothesis was that there was no association between RFC1 gene polymorphism (rs1051266) and non-syndromic cleft lip and palate in the South Indian population. The study aimed to determine the association between RFC1 gene polymorphism (rs1051266) and non-syndromic cleft lip and palate in the South Indian population.
MATERIALS AND METHODS
This case-control study was conducted in the Department of Orthodontics of our institution. The study was approved by the scientific review board of our university with reference number SRB/SDC/FACULTY/22/ORTHO/052 and ethical clearance was obtained with reference number IHEC/SDC/FACULTY/22/ORTHO/58. Patients reporting with cleft lip and/or palate to the cleft and craniofacial unit of our university were screened. Patients with NSCL/P without associated syndromes or familial history of cleft lip and palate with ages ranging from 1 year to 17 years consisting of both genders belonging to the South Indian population were included in the study.
The exclusion criteria included patients with habits such as smoking, diabetes, pan chewing, tobacco chewing, oral lesions such as submucous fibrosis, erythroplakia, leukoplakia, oral submucous fibrosis, diabetic patients, and patients with systemic diseases. Children whose parents had the above habits or diseases were excluded from the study.
The sample consisted of two groups. Group 1 was the study group with subjects having non-syndromic cleft lip and palate patients while group 2 was the control group with patients without cleft lip and palate malformations. Twenty-five patients were included in each group. Informed consent was obtained from all the adult patients and the parents of the children who were included in the study. Five milliliters of venous blood was drawn from the antecubital fossa of the patient and collected in sterile tubes with a small amount of ethylenediaminetetraacetic acid. The blood was carefully blended to prevent clot formation (22). RFC1 gene polymorphism (rs1051266) was assessed using polymerase chain reaction (PCR) amplification and restriction digestion. Amplification of DNA around the (rs1051266) polymorphic site of the RFC1 gene was performed. The forward primer was: 5’- AGCGTCACCTTCGTCCC - 3’ and the reverse primer was 5’- TCCCGCGTGAAGTTCTTG -3’. The amplification of DNA was performed in 20 µL volumes using 10 ng of genomic DNA, 5 pmol/µL, each of the forward and reverse primers along with PCR Master Mix (Takara, Shiga, Japan). The cycling conditions consisted of an initial denaturation at 94°C for 5 minutes, denaturation at 94°C for 35 seconds, annealing at 60°C for 35 seconds, extension at 72°C for 35 seconds, and a final extension at 72°C for 5 minutes. A 5 µL volume of PCR product was checked on a 1% agarose gel, and 15 µL of PCR product was digested using a restriction enzyme AseI (New England Biolabs, Hitchin, UK). Digestion was carried out at 37°C for 2 hours. The digested product was visualized on 2% agarose gel. After the blood samples were obtained from both groups, and their genomic DNA was put through the PCR RFLP test, the Hha1 enzyme was used to cleave the PCR product, and different band patterns were obtained.
The band patterns were run along a standard DNA ladder. The frequency of homozygous alleles; GG and AA and heterozygous allele GA was determined.
STATISTICAL ANALYSIS
Statistical analysis was done with SPSS version 22.0. The Hardy-Weinberg equilibrium (HWE) was tested in each group using the Chi-Square test. Allele ratio and genotype distribution of NSCL/P patients and healthy controls were evaluated. The odds ratio (OR) with 95% confidence intervals was used to calculate the risk related to specific alleles or genotypes. A p-value ≤ 0.05 was considered to be statistically significant.
RESULTS
A total of 50 patients, 25 each in group 1 and group 2 were evaluated respectively. Patients in group 1 and group 2 had a mean age of 4.99±5.2 years and 23.24±6.8 years, respectively. The ancestral allele associated with RFC1 gene polymorphism (rs1051266) was the G allele and the variant allele was the A allele. (https://asia.ensembl.org/Homo_sapiens/Variation/Explore?db=core;r=1:11795821-11796821;v=rs1801133;vdb=variation;vf=122492).
The agarose gel electrophoretogram showed amplification with an amplicon size of 230 bp around the polymorphic site RFC1 gene polymorphism (rs1051266) (Figure 1). Agarose gel electrophoretogram of the HhaI digested amplicon of RFC1 gene at (rs1051266) depicted the homozygous GG genotype when three bands were present at 125, 68 and 37 base pairs; heterozygous genotype GA when bands were present at 162, 125, 68 and 37 base pairs; and homozygous genotype AA when two bands were present at 162 and 68 base pairs. These were compared with a standard DNA ladder (Lane M=100 base pair DNA marker) (Figure 2). The frequency of the GG genotype was 12% (3 out of 25), the GA genotype was 60% (15 out of 25) and the AA genotype was 28% (7 out of 25)in group 1 (case group) (Table 1). The genotype frequency in group 2 (control group) was found to be GG=28%, GA=52%, and AA =20%. The G allele frequency was 0.42 and 0.54 in group 1 and group 2 respectively (Table 1). The A allele frequency was 0.58 and 0.46 in group 1 and group 2, respectively. There was no statistically significant deviation from Hardy Weinberg Equilibrium with a p-value of <0.247 and <0.815 in the case group and control group respectively (Table 1).
The frequency distribution of the gene polymorphism between the two groups did not yield any statistically significant results. The frequency distribution of the dominant genotype GG compared to the other genotype GA and AA had an odd ratio of 0.3506 (0.0791-1.5544) and a p-value of 0.1678 The frequency distribution of the recessive genotype AA compared to the other genotype GA and GG had an odd ratio of1.5556 (0.4187-5.7795) and P value of 0.5094. The distribution of G allele and A allele between the two groups had an odd ratio of 0.6169 [0.2798 - 1.3597] and p p-value of 0.2309. Since the Odd’s ratio has a range of less than 1 to greater than 1 for both the dominant and recessive genotypes, it is difficult to ascertain its protective or destructive nature. Classification of the genotypes based on genetic models such as dominant, recessive, or additive (allelic) did not present any significant association of the polymorphism marker with the disease status (Table 2). The null hypothesis was accepted.
The comparison of allele frequencies revealed the distribution of G and A alleles of subjects belonging to the South Indian population in the present study matched the frequency distribution of the American, European, Asian, and Global populations (Figure 3). The frequency of the ancestral G allele was less in the African population (Figure 3).

Figure 1. Agarose gel electrophoretogram showing partial amplification of RFC1 gene spanning polymorphic site (rs1051266) run along with standard DNA ladder (Lane M=100 bp DNA marker).

Figure 2. Agarose gel electrophoretogram showing HhaI digested amplicon of RFC1 gene at (rs1051266) (Homozygous: GG - 125+68+37 bp; Heterozygous: GA-162+125+68+37 bp; Homozygous: AA-162+68 bp) (Lane M=100 bp DNA marker).
Table 1. Genotype frequencies of RFC1 gene polymorphism (rs1051266) among the cases and controls.
|
Groups |
GG |
GA |
AA |
G |
A |
HWE (p-value)* |
|
Case (N=25) |
3 |
15 |
7 |
0.42 |
0.58 |
0.247 |
|
Control (N=25) |
7 |
13 |
5 |
0.54 |
0.46 |
0.815 |
*For departure from Hardy-Weinberg equilibrium (HWE), chi-square with one degree of freedom. The genotype frequency of cases and controls do not differ significantly χ 2df (P =0.3541).
P value ≤ 0.05 is considered statistically significant.
Table 2. Overall genotype distribution of the RFC1 gene polymorphism (rs1051266) in cases and controls.
|
Dominant |
||||
|
Genotypes |
Case |
Control |
Unadjusted OR (95% CI) |
P value |
|
GG |
3 |
7 |
0.3506 (0.0791 - 1.5544) |
0.1678 |
|
GA + AA |
22 |
18 |
||
|
Recessive |
||||
|
AA |
7 |
5 |
1.5556 (0.4187 - 5.7795) |
0.5094 |
|
GG + GA |
18 |
20 |
||
|
Allele |
||||
|
G |
21 |
27 |
0.6169 (0.2798 - 1.3597) |
0.2309 |
|
A |
29 |
23 |
||
P value ≤ 0.05 is considered statistically significant.
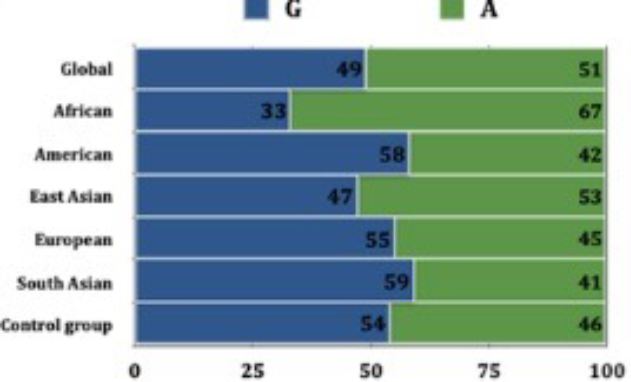
Figure 3. The graph depicts the allele frequency of RFC1 gene (rs1051266) polymorphism in various study population compared to the present study group (Ensembl database).
DISCUSSION
Folate is necessary for several biological functions including the production and maintenance of DNA. RFC1 plays an important role in maintaining folate levels by coding for the protein involved in the entry of folate into the cell.
Polymorphisms of the RFC1 gene may affect folate transportation into the cell. Adequate folate levels are required for proper development of the embryo during pregnancy. Gene polymorphisms and mutations of the RFC1 gene have been associated with the development of non-syndromic cleft lip and palate in humans. The proper development of facial structures during embryonic development can be disrupted by mutations in these genes.
There is conflicting evidence regarding the association between allelic RFC1 A80G polymorphism and NSCL/P in diverse populations (23-27). Existing reports suggest that RFC1A80G polymorphism is associated with NSCL/P in the East Indian population (26). However, the present study did not show such an association. The earlier study (26) was conducted in the Eastern Indian population with Caucasian samples whereas the present study was conducted on the South Indian population consisting of subjects belonging to the Dravidian race. A previous study conducted on the South Indian population 10 years ago showed an association between dominant and allelic models of the RFC1 gene and NSCL/P (20). RFC1 gene polymorphism RFC1 A80G polymorphism was associated with the NSCL/P in the Iranian population as well (28). A recent systematic review showed that the GG genotype was associated with NSCL/P. This was more prevalent in the Caucasian population compared to other ethnic groups (29). In the current study, the prevalence of the GG genotype was less in patients with NSCL/P compared to the GA and AA genotypes. RFC1 gene polymorphism was associated with NSCL/P in the Chinese population and the A allele had a greater risk of the development of the anomaly (30). The RFC1 gene polymorphism affects other organs, especially the heart, and is associated with congenital heart defects in the Chinese and American population (31, 32).
It is important to remember that cleft lip and palate are frequently multifactorial and may be due to a combination of genetic and environmental factors. Multidisciplinary management has been advocated for the treatment of non-syndromic cleft lip and palate with surgery to repair the nose, alveolar bone, and lip, orthodontic treatment to correct the teeth, speech therapy, psychological counseling, and pediatric dentistry (33, 34). Facial esthetics in cleft lip and palate patients may be improved with the systematic assessment of the mid-face and mandible (35). Some of the recent advances in the management of NSCL/P are the role of stem cell therapy to regenerate bone at the cleft site (36) and the use of artificial intelligence to plan treatment (37).
Future studies with a larger sample size may be contemplated to confirm the results of the present study.
CONCLUSION
The frequency of the GG genotype was 12% and 28%, the GA genotype was 60% and 52% and the AA genotype was 28% and 20% in group 1 (case group) and group 2 (control group) respectively The A allele frequency was 0.58 and 0.46 in group 1 and group 2, respectively. There was no statistically significant deviation from the Hardy-Weinberg Equilibrium. The Odd’s ratio has a range of less than 1 to greater than 1 for both the dominant GG genotype and recessive AA genotype making it difficult to determine its protective or destructive effect. The dominant and recessive genotypes and the alleles did not present any significant association of the polymorphism marker with NSCLP/P. From the above study, the RFC1 gene (rs1051266) polymorphism does not have a significant association in the development of non-syndromic cleft lip and palate in the South Indian population. However, larger studies are required to confirm these results.
AUTHOR CONTRIBUTION STATEMENT: Conceptualization and design: A.S.F. and V.P.J.; Literature review: A.S.F. and R.M.J.; Methodology and validation: A.S.F. and V.P.J.; Formal analysis: A.S.F. and V.P.J.; Investigation and data collection: A.S.F.; Resources: R.M.J.; Data analysis and interpretation: A.S.F. and V.P.J.; Writing-original draft preparation: A.S.F. and R.M.J.; Writing-review & editin: A.S.F. and V.P.J.; Funding acquisition: R.M.J.
REFERENCES
1. Hammond N.L., Dixon M.J. Revisiting the embryogenesis of lip and palate development. Oral Dis. 2022; 28 (5): 1306-1326.
2. Vyas T., Gupta P., Kumar S., Gupta R., Gupta T., Singh H.P. Cleft of lip and palate: A review. J Family Med Prim Care. 2020; 9 (6): 2621-2625.
3. Kosowski T.R., Weathers W.M., Wolfswinkel E.M., Ridgway E.B. Cleft palate. Semin Plast Surg. 2012; 26 (4): 164-169.
4. DeRoo L.A., Wilcox A.J., Drevon C.A., Lie R.T. First-trimester maternal alcohol consumption and the risk of infant oral clefts in Norway: a population-based case-control study. Am J Epidemiol. 2008; 168 (6): 638-646.
5. Sweeney E., Fryer A. Nasomaxillary hypoplasia and severe orofacial clefting in a child of a mother with phenylketonuria. J Inherit Metab Dis. 2002; 25 (1): 77-79.
6. Peterka M., Tvrdek M., Likovský Z., Peterková R., Fára M. Maternal hyperthermia and infection as one of possible causes of orofacial clefts. Acta Chir Plast. 1994; 36 (4): 114-118.
7. Singh R., Kumar N., Arora S., Bhandari R., Jain A. Fetal hydantoin syndrome and its anaesthetic implications: a case report. Case Rep Anesthesiol. 2012; 2012: 370412.
8. Nichols M.M. Fetal anomalies following maternal trimethadione ingestion. J Pediatr. 1973; 82 (5): 885-886.
9. Hyoun S.C., Običan S.G., Scialli A.R. Teratogen update: methotrexate. Birth Defects Res A Clin Mol Teratol. 2012; 94 (4): 187-207.
10. Sicinska E., Kubiak K., Madej D., Granda D., Kaluza J. Main sources and predictive factors of folate intake in female university students. Nutrition. 2024; 120: 112359.
11. Stamm R.A., Houghton L.A. Nutrient intake values for folate during pregnancy and lactation vary widely around the world. Nutrients. 2013; 5 (10): 3920-3947.
12. Greenberg J.A., Bell S.J., Guan Y., Yu Y.H. Folic Acid supplementation and pregnancy: more than just neural tube defect prevention. Rev Obstet Gynecol. 2011; 4 (2): 52-59.
13. Irwin R.E., Pentieva K., Cassidy T., et al. The interplay between DNA methylation, folate and neurocognitive development. Epigenomics. 2016; 8 (6): 863-879.
14. Au K.S., Findley T.O., Northrup H. Finding the genetic mechanisms of folate deficiency and neural tube defects-Leaving no stone unturned. Am J Med Genet A. 2017; 173 (11): 3042-3057.
15. Dixon M.J., Marazita M.L., Beaty T.H., Murray J.C. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011; 12 (3): 167-178.
16. Deshpande A.S., Goudy S.L. Cellular and molecular mechanisms of cleft palate development. Laryngoscope Investig Otolaryngol. 2018; 4 (1): 160-164.
17. Yu Y., Zuo X., He M., et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun. 2017; 8: 14364.
18. Matherly L.H., Goldman D.I. Membrane transport of folates. Vitam Horm. 2003; 66: 403-456.
19. Sirotnak F.M., Tolner B. Carrier-mediated membrane transport of folates in mammalian cells. Annu Rev Nutr. 1999;19: 91-122.
20. Lakkakula B., Murthy J., Gurramkonda V.B. Relationship between reduced folate carrier gene polymorphism and non-syndromic cleft lip and palate in Indian population. J Matern Fetal Neonatal Med. 2015; 28 (3): 329-332.
21. Mossey P.A., Little J., Munger R.G., Dixon M.J., Shaw W.C. Cleft lip and palate. Lancet. 2009; 374 (9703): 1773-1785.
22. Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988; 16 (3): 1215.
23. Sun M., Yuan C., Chen J., et al. Association between RFC1 A80G polymorphism and the susceptibility to nonsyndromic cleft lip with or without cleft palate: a meta-analysis. Ann Transl Med. 2019; 7 (23): 721.
24. Bezerra J.F., Oliveira G.H., Soares C.D., et al. Genetic and non-genetic factors that increase the risk of non-syndromic cleft lip and/or palate development. Oral Dis. 2015; 21(3): 393-399.
25. Girardi A., Martinelli M., Cura F., et al. RFC1 and non-syndromic cleft lip with or without cleft palate: an association based study in Italy. J Craniomaxillofac Surg. 2014; 42 (7): 1503-1505.
26. Kumari P., Ali A., Sukla K.K., Singh S.K., Raman R. Lower incidence of nonsyndromic cleft lip with or without cleft palate in females: is homocysteine a factor?. J Biosci. 2013; 38 (1): 21-26.
27. Bufalino A., Ribeiro Paranaíba L.M., Nascimento de Aquino S., Martelli-Júnior H., Oliveira Swerts M.S., Coletta R.D. Maternal polymorphisms in folic acid metabolic genes are associated with nonsyndromic cleft lip and/or palate in the Brazilian population. Birth Defects Res A Clin Mol Teratol. 2010; 88 (11): 980-986.
28. Soghani B., Ebadifar A., Khorram Khorshid H.R., Kamali K., Hamedi R., Aghakhani Moghadam F. The study of association between reduced folate carrier 1 (RFC1) polymorphism and non-syndromic cleft lip/palate in Iranian population. Bioimpacts. 2017; 7 (4): 263-268.
29. Imani M.M., Mozaffari H.R., Sharifi R., Sadeghi M. Polymorphism of reduced folate carrier 1 (A80G) and non-syndromic cleft lip/palate: A systematic review and meta-analysis. Arch Oral Biol. 2019; 98: 273-279.
30. Wang Y., Song X., Guo J., Zhu W. Relationship between genetic polymorphisms of RFC1 A80G and nonsymdromic cleft lip with or without palate. Journal of hygiene research. 2009; 38 (3): 276-9.
31. Pei L., Zhu H., Zhu J., Ren A., Finnell R.H., Li Z. Genetic variation of infant reduced folate carrier (A80G) and risk of orofacial defects and congenital heart defects in China. Ann Epidemiol. 2006; 16 (5): 352-356.
32. Shaw G.M., Zhu H., Lammer E.J., Yang W., Finnell R.H. Genetic variation of infant reduced folate carrier (A80G) and risk of orofacial and conotruncal heart defects. Am J Epidemiol. 2003; 158 (8): 747-752.
33. Natarajan P.G., Mahipathy S.R.R.V., Nalabothu P. Anatomic Subunit Cleft Lip Repair: New Repair Concept Based on Nostril Sill and Philtrum-White Roll Junction. J Craniofac Surg. Published online September 4, 2024.
34. Alkadhi O.H., Alotaibi L.H., Alrashoud R.R., Almutairi M.H., Al Matar H.A., Mallineni S.K. Effect of Maxillary Expansion on the Maxillary Arch Width in Patients with Bilateral Cleft Palate: A Review. Children (Basel). 2023; 10 (5): 762.
35. Anishya D., Nagesh S. Assessment of Nasal Aesthetic Parameters in Patients with Unilateral Cleft Lip and Palate - A Retrospective Study. Cleft Palate Craniofac J. Published online June 11, 2024.
36. Jaber M., Alshikh Ali A.M., El Saleh R.I., Prasad P. The Use of Stem Cells in Bone Regeneration of Cleft Lip and Palate Patients: A Systematic Review. J Clin Med. 2024;13 (17): 5315.
37. Tovani-Palone MR. Management of orofacial clefts in times of artificial intelligence: advances and challenges. Eur Arch Paediatr Dent. 2024; 25 (5): 773-774.
 Odovtos -Int J Dent Sc endoses to CC-BY-NC-SA 4.0.
Odovtos -Int J Dent Sc endoses to CC-BY-NC-SA 4.0.