Abstract
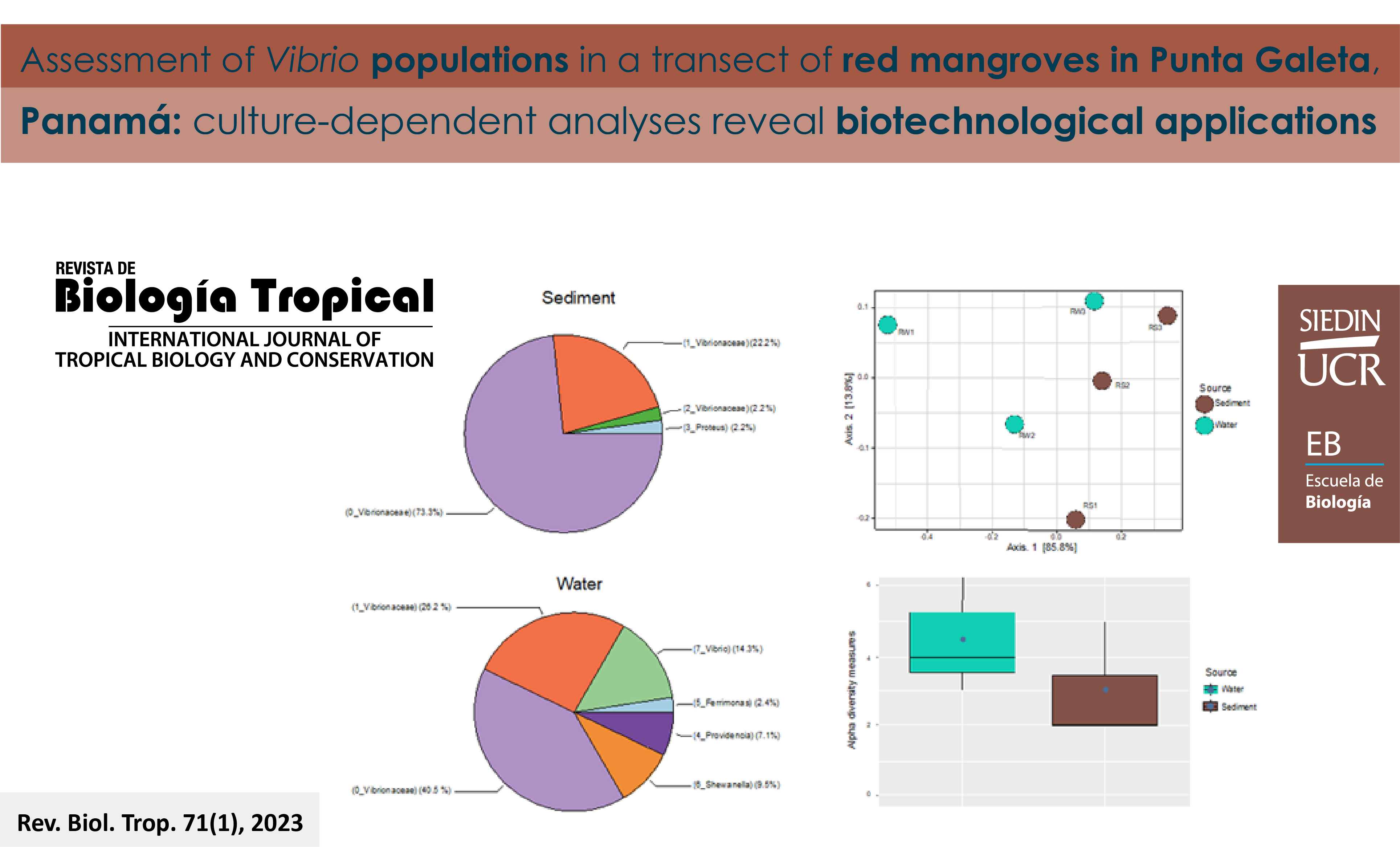
Introduction: Rhizophora mangle is considered a niche for microorganisms with potentially novel and complex degrading enzymes. Objective: To characterize Vibrio populations using culture-dependent methods, from samples collected from sediments and water along a red mangrove transect composed of three sites. Methods: Strains were characterized according to their distribution, diversity, degradation of organic matter, and environmental parameters. Results: Bacterial densities were strongly associated with temperature and salinity. A total of 87 good-quality sequences representing the isolates from the three sites, were binned into eight OTUs (Operational taxonomic units). Taxonomic assignment indicated that the dominant members were Vibrionaceae. Beta diversity analyses showed that bacterial communities clustered by sample source rather than spatial distribution, and that alpha diversity was found to be higher in water than in sediment. Three percent of the strains from water samples could degrade carboxyl-methyl cellulose with the smallest enzymatic indexes compared to 4 % of the strains from sediment samples that showed the highest enzymatic indexes. Two strains identified as Vibrio agarivorans degraded cellulose and agarose, producing the highest enzymatic indexes. Conclusions: We found higher bacterial densities and diversity in the bacterial communities of the water samples compared to the sediment, with different OTUs including those similar to Ferrimonas, Providencia, or Shewanella which were not isolated in the sediment. Vibrio OTUs were shown to degrade cellulose in both sample types. The results of this study highlight the importance of red mangroves as Vibrio habitats and as reservoirs of potential enzyme sources with biotechnological applications.
References
Archambault, P., & Bourget, E. (1996). Scales of coastal heterogeneity and benthic intertidal species richness, diversity and abundance. Marine Ecology Progress Series, 136, 111–121. https://doi:10.3354/meps136111
Bibi, F., Ullah, I., Alvi, S. A., Bakhsh, S. A., Yasir, M., Al-Ghamdi, A. A. K., & Azhar, E. I. (2017). Isolation, diversity, and biotechnological potential of rhizo- and endophytic bacteria associated with mangrove plants from Saudi Arabia. Genetics and Molecular Research, 16(2), gmr16029657. https://doi.org/10.4238/gmr16029657
Blackwell, K. D., & Oliver, J. D. (2008). The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. Journal of Microbiology, 46(2), 146–153. https://doi.org/10.1007/s12275-007-0216-2
Blanco-Abad, V., Ansede-Bermejo, J., Rodriguez-Castro, A., & Martinez-Urtaza, J. (2009). Evaluation of different procedures for the optimized detection of Vibrio parahaemolyticus in mussels and environmental samples. International Journal of Food Microbiology, 129(3), 229–236. https://doi.org/10.1016/j.ijfoodmicro.2008.11.028
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Peña, A. G., Goodrich, J. K., Gordon, J. I., Huttley, G. A., Kelley, S. T., Knights, D., Koenig, J. E., Ley, R. E., Lozupone, C. A., McDonald, D., Muegge, B. D., Pirrung, M., … & Knight, R. (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5), 335–336. https://doi.org/10.1038/nmeth.f.303
Chao, A. (1987). Estimating the population size for capture-recapture data with unequal catchability. Biometrics, 43(4), 783–791.
De Vries, A., & Ripley, B. D. (2016). ggdendro: Create Dendrograms and Tree Diagrams Using 'ggplot2' (Version 0.1.20). https://github.com/andrie/ggdendro.
DeYoe, H., Lonard, R. I., Judd, F. W., Stalter, R., & Feller, I. (2020). Biological Flora of the Tropical and Subtropical Intertidal Zone: Literature Review for Rhizophora mangle L. Journal of Coastal Research, 36(4), 857–884. https://doi.org/10.2112/JCOASTRES-D-19-00088.1
Dhariwal, A., Chong, J., Habib, S., King, I. L., Agellon, L. B., & Xia, J. (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Research, 45(W1), W180–W188. https://doi.org/10.1093/nar/gkx295
Dias, A. C., Andreote, F. D., Dini-Andreote, F., Lacava, P. T., Sá, A. L., Melo, I. S., Azevedo, J. L., & Araújo, W. L. (2009). Diversity and biotechnological potential of culturable bacteria from Brazilian mangrove sediment. World Journal of Microbiology and Biotechnology, 25(7), 1305–1311. https://doi.org/10.1007/s11274-009-0013-7
Dias, A. C., Taketani, R. G., Andreote, F. D., Luvizotto, D. M., da Silva, J. L., Nascimento, R., & de Melo, I. S. (2012). Interspecific variation of the bacterial community structure in the phyllosphere of the three major plant components of mangrove forests. Brazilian Journal of Microbiology, 43(2), 653–660. https://doi.org/10.1590/S1517-83822012000200030
Drzewiecka, D. (2016). Significance and Roles of Proteus spp. Bacteria in Natural Environments. Microbial Ecology, 72(4), 741–758. https://doi.org/10.1007/s00248-015-0720-6
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27(16), 2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Ellison, A., Farnsworth, E., & Moore, G. (2015). Rhizophora mangle. The IUCN Red List of Threatened Species 2015: e.T178851A69024847. IUCN https://dx.doi.org/10.2305/IUCN.UK.2015-1.RLTS.T178851A69024847.en.
Florencio, C., Couri, S., & Farinas, C. S. (2012). Correlation between Agar Plate Screening and Solid-State Fermentation for the Prediction of Cellulase Production by Trichoderma Strains. Enzyme Research, 2012, 793708, 1-7. https://doi.org/10.1155/2012/793708
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., & Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Applied and Environmental Microbiology, 74(8), 2461–2470. https://doi.org/10.1128/AEM.02272-07
Gomes, N. C., Cleary, D. F., Pinto, F. N., Egas, C., Almeida, A., Cunha, A., Mendonça-Hagler, L. C., & Smalla, K. (2010). Taking root: enduring effect of rhizosphere bacterial colonization in mangroves. PLOS ONE, 5(11), e14065. https://doi.org/10.1371/journal.pone.0014065
Gomes, N., Cleary, D., Pires, A., Almeida, A., Cunha, A., Mendonça-Hagler, L., & Smalla, K. (2014). Assessing variation in bacterial composition between the rhizospheres of two mangrove tree species. Estuarine, Coastal and Shelf Science, 139, 40–45. https://doi.org/10.1016/j.ecss.2013.12.022
Gonzalez, A., King, A., Robeson II, M. S., Song, S., Shade, A., Metcalf, J. L., & Knight, R. (2012). Characterizing microbial communities through space and time. Current Opinion in Biotechnology, 23(3), 431–436. https://doi.org/10.1016/j.copbio.2011.11.017
Gonzalez-Acosta, B., Bashan, Y., Hernandez-Saavedra, N. Y., Ascencio, F., & Cruz-Agüero, G. (2006). Seasonal seawater temperature as the major determinant for populations of culturable bacteria in the sediments of an intact mangrove in an arid region. FEMS Microbiology Ecology, 55(2), 311–321. https://doi.org/10.1111/j.1574-6941.2005.00019.x
Hamzah, T. N. T., Lee, S. Y., Hidayat, A., Terhem, R., Faridah-Hanum, I., & Mohamed, R. (2018). Diversity and characterization of endophytic fungi isolated from the tropical mangrove species, Rhizophora mucronata, and identification of potential antagonists against the soil-borne fungus, Fusarium solani. Frontiers in Microbiology, 9, 1707. https://doi.org/10.3389/fmicb.2018.01707
Holguin, G., Gonzalez-Zamorano, P., de-Bashan, L. E., Mendoza, R., Amador, E., & Bashan, Y. (2006). Mangrove health in an arid environment encroached by urban development - a case study. The Science of the Total Environment, 363(1–3), 260–274. https://doi.org/10.1016/j.scitotenv.2005.05.026
Hu, Z., Lin, B. K., Xu, Y., Zhong, M. Q., & Liu, G. M. (2009). Production and purification of agarase from a marine agarolytic bacterium Agarivorans sp. HZ105. Journal of Applied Microbiology, 106(1), 181–190. https://doi.org/10.1111/j.1365-2672.2008.03990.x
Jacobs, J. M., Rhodes, M., Brown, C. W., Hood, R. R., Leight, A., Long, W., & Wood, R. (2014). Modeling and forecasting the distribution of Vibrio vulnificus in Chesapeake Bay. Journal of Applied Microbiology, 117(5), 1312–1327. https://doi.org/10.1111/jam.12624
Janelidze, N., Jaiani, E., Lashkhi, N., Tskhvediani, A., Kokashvili, T., Gvarishvili, T., Jgenti, D., Mikashavidze, E., Diasamidze, R., Narodny, S., Obiso, R., Whitehouse, C. A., Huq, A., & Tediashvili, M. (2011). Microbial water quality of the Georgian coastal zone of the Black Sea. Marine Pollution Bulletin, 62(3), 573–580. https://doi.org/10.1016/j.marpolbul.2010.11.027
Johnson, C. N. (2015). Influence of environmental factors on Vibrio spp. in coastal ecosystems. Microbiology Spectrum, 3(3), 1–18. https://doi.org/10.1128/microbiolspec.VE-0008-2014
Liu, G., Wu, S., Jin, W., & Sun, C. (2016). Amy63, a novel type of marine bacterial multifunctional enzyme possessing amylase, agarase and carrageenase activities. Scientific Reports, 6, 18726. https://doi.org/10.1038/srep18726
Lüdeke, C. H., Gonzalez-Escalona, N., Fischer, M., & Jones, J. L. (2015). Examination of clinical and environmental Vibrio parahaemolyticus isolates by multi-locus sequence typing (MLST) and multiple-locus variable-number tandem-repeat analysis (MLVA). Frontiers in Microbiology, 6, 564. https://doi.org/10.3389/fmicb.2015.00564
Macián, M. C., Ludwig, W., Schleifer, K. H., Pujalte, M. J., & Garay, E. (2001). Vibrio agarivorans sp. nov., a novel agarolytic marine bacterium. International Journal of Systematic and Evolutionary Microbiology, 51(6), 2031–2036. https://doi.org/10.1099/00207713-51-6-2031
Mackey, K., Hunter-Cevera, K., Britten, G. L., Murphy, L. G., Sogin, M. L., & Huber, J. A. (2017). Seasonal Succession and Spatial Patterns of Synechococcus Microdiversity in a Salt Marsh Estuary Revealed through 16S rRNA Gene Oligotyping. Frontiers in Microbiology, 8, 1496. https://doi.org/10.3389/fmicb.2017.01496
McDonald, D., Clemente, J. C., Kuczynski, J., Rideout, J. R., Stombaugh, J., Wendel, D., Wilke, A., Huse, S., Hufnagle, J., Meyer, F., Knight, R., & Caporaso, J. G. (2012). The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. GigaScience, 1(1), 7. https://doi.org/10.1186/2047-217X-1-7
McMurdie, P. J., & Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE, 8(4), e61217. https://doi.org/10.1371/journal.pone.0061217
Müllner, D. (2013). fastcluster: Fast Hierarchical, Agglomerative Clustering Routines for R and Python. Journal of Statistical Software, 53(9), 1–18. https://doi.org/10.18637/jss.v053.i09
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., Minchin, P. R., O’Hara, R. B., Simpson, G. L., Solymos, P., Stevens, M. H. H., Szöcs, E., & Wagner, H. (2020). Vegan: Community ecology package. Ordination methods, diversity analysis and other functions for community and vegetation ecologists (Version 2.5). https://CRAN. R-project. org/package= vegan
Pfeffer, C. S., Hite, M. F., & Oliver, J. D. (2003). Ecology of Vibrio vulnificus in estuarine waters of eastern North Carolina. Applied and Environmental Microbiology, 69(6), 3526–3531. https://doi.org/10.1128/AEM.69.6.3526-3531.2003
Pruesse, E., Peplies, J., & Glöckner, F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics, 28(14), 1823–1829. https://doi.org/10.1093/bioinformatics/bts252
Pruzzo, C., Huq, A., Colwell, R. R., & Donelli, G. (2005). Pathogenic Vibrio species in the marine and estuarine environment. In S. Belkin, & R. R. Colwell (Eds.), Ocean and Health Pathogens in the Marine Environment (pp. 217–252). Springer-Verlag.
Rameshkumar, N., & Nair, S. (2009). Isolation and molecular characterization of genetically diverse antagonistic, diazotrophic red-pigmented vibrios from different mangrove rhizospheres. FEMS Microbiology Ecology, 67(3), 455–467. https://doi.org/10.1111/j.1574-6941.2008.00638
Robinson, J. D., Diaz-Ferguson, E., Poelchau, M. F., Pennings, S., Bishop, T. D., & Wares, J. (2010). Multiscale diversity in the marshes of the Georgia coastal ecosystems LTER. Estuaries and Coasts, 33(4), 865–877. https://doi.org/10.1007/s12237-009-9188-2
Rocha, L. L., Colares, G. B., Nogueira, V. L., Paes, F. A., & Melo, V. M. (2016). Distinct Habitats Select Particular Bacterial Communities in Mangrove Sediments. International Journal of Microbiology, 2016, 3435809, 1–6. https://doi.org/10.1155/2016/3435809
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B., Thallinger, G. G., Van Horn, D. J., & Weber, C. F. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75(23), 7537–7541. https://doi.org/10.1128/AEM.01541-09
Soares Júnior, F. L., Dias, A. C., Fasanella, C. C., Taketani, R. G., de Souza Lima, A. O., Melo, I. S., & Andreote, F. D. (2014). Endo- and exoglucanase activities in bacteria from mangrove sediment. Brazilian Journal of Microbiology, 44(3), 969–976. https://doi.org/10.1590/s1517-83822013000300048
Sousa, W. P., & Mitchell, B. J. (1999). The Effect of Seed Predators on Plant Distributions: Is There a General Pattern in Mangroves? Oikos, 86(1), 55–66. https://doi.org/10.2307/3546569
Sá, A. L., Dias, A. C., Quecine, M. C., Cotta, S. R., Fasanella, C. C., Andreote, F. D., & de Melo, I. S. (2014). Screening of endoglucanase-producing bacteria in the saline rhizosphere of Rhizophora mangle. Brazilian Journal of Microbiology, 45(1), 193–197. https://doi.org/10.1590/s1517-83822014000100025
Tagliavia, M., Salamone, M., Bennici, C., Quatrini, P., & Cuttitta, A. (2019). A modified culture medium for improved isolation of marine vibrios. MicrobiologyOpen, 8(9), e00835. https://doi.org/10.1002/mbo3.835
Takemura, A. F., Chien, D. M., & Polz, M. F. (2014). Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Frontiers in Microbiology, 5, 38. https://doi.org/10.3389/fmicb.2014.00038
Teather, R. M., & Wood, P. J. (1982). Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Applied and Environmental Microbiology, 43(4), 777–780. https://doi.org/10.1128/aem.43.4.777-780.1982
Thatoi, H., Behera, B. C., Mishra, R. R., & Dutta, S. K. (2013). Biodiversity and biotechnological potential of microorganisms from mangrove ecosystems: a review. Annals of Microbiology, 63(1), 1–19. https://doi.org/10.1007/s13213-012-0442-7
Thompson, F. L., Iida, T., & Swings, J. (2004). Biodiversity of vibrios. Microbiology and Molecular Biology Reviews, 68(3), 403–431. https://doi.org/10.1128/MMBR.68.3.403-431.2004
Turner, J. W., Good, B., Cole, D., & Lipp, E. K. (2009). Plankton composition and environmental factors contribute to Vibrio seasonality. The ISME Journal, 3(9), 1082–1092. https://doi.org/10.1038/ismej.2009.50
Wang, Q., Garrity, G. M., Tiedje, J. M., & Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73(16), 5261–5267. https://doi.org/10.1128/AEM.00062-07
Wetz, J. J., Blackwood, A. D., Fries, J. S., Williams, Z. F., & Noble, R. T. (2008). Trends in total Vibrio spp. and Vibrio vulnificus concentrations in the eutrophic Neuse River Estuary, North Carolina, during storm events. Aquatic Microbial Ecology, 53(1), 141–149. https://doi.org/10.3354/ame0122
Wickham, H. (2007). Reshaping Data with the reshape Package. Journal of Statistical Software, 21(12), 1–20. https://doi.org/10.18637/jss.v021.i12
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. https://ggplot2.tidyverse.org
Wilke, C. (2017). Package cowplot: Streamlined Plot Theme and Plot Annotations for ggplot2. https://wilkelab.org/cowplot/
Zhang, Y., Yang, Q., Ling, J., Van Nostrand, J. D., Shi, Z., Zhou, J., & Dong, J. (2017). Diversity and Structure of Diazotrophic Communities in Mangrove Rhizosphere, Revealed by High-Throughput Sequencing. Frontiers in Microbiology, 8, 2032. https://doi.org/10.3389/fmicb.2017.02032
Zhu, D. H., Song, Q. L., Nie, F. H., Wei, W., Chen, M. M., Zhang, M., Lin, H. Y., Kang, D. J., Chen, Z. B., Hay, A. G., & Chen, J. J. (2022). Effects of environmental and spatial variables on bacteria in Zhanjiang mangrove sediments. Current Microbiology, 79(4), 97. https://doi.org/10.1007/s00284-022-02774-z
##plugins.facebook.comentarios##

This work is licensed under a Creative Commons Attribution 4.0 International License.
Copyright (c) 2023 Revista de Biología Tropical